Lightweight U-Nets
Low-Memory CNNs Enabling Real-Time Ultrasound Segmentation Towards Mobile Deployment
Sagar Vaze, Weidi Xie, Ana Namburete
Paper Code
We propose methods to build CNNs capable of running at 30 frames per second on a CPU and of having small memory footprints, facilitating deployment on mobile devices. Our work is catered towards ultrasound imaging, whose high frame rates require fast image analysis, and whose portable nature benefits form analysis algorithms capable of running on mobile devices.
Context
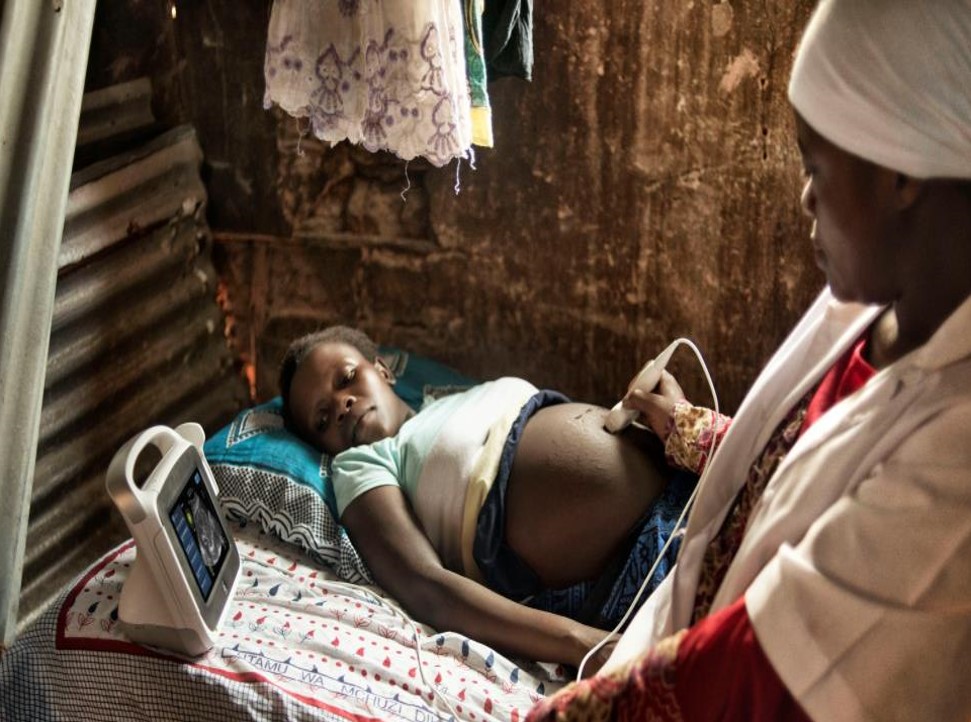
Ultrasound imaging is still widely used around the world despite the presence of higher resolution modalities such as MRI and CT. It
is often favoured due to its low cost, portable nature and high frame rates, with the market now containing a number of hand-held
probes capable of imaging in real-time for a few hundred dollars.
For these reasons, ultrasound is used in the imaging of moving fetuses and is useful more generally in the developing world (particularly in low-income,
rural areas). Therefore, image analysis techniques which can operate efficiently on a mobile device have high utility. However, the state-of-the-art
medical image analysis algorithms, namely convolutional neural networks (CNNs), come with high computational costs. CNNs typically require millions of
parameters to define, the storage of which requires significant memory and hinders deployment on mobiles. In addition, each CNN parameter necessitates a
number of multiplications and additions during the model's computation, resulting in lengthy run-times.
The problem we tackle in this work is that of efficient neural networks for application to ultrasound imaging, with the ultimate goal being the development
of imaging algorithms which are capable of running in real-time on a mobile device, to deliver automated ultrasound analysis at point-of-care, alongside the imaging
itself.
Technical Contributions
We tackle the image analysis task of semantic segmentation, for which the CNN architectures used typically have the largest number of
parameters and require the longest inference times. For instance, the U-Net, the most commonly
applied segmentation network in the medical imaging literature, requires 130 million parameters to define, taking nearly half a gigabyte in memory.
We propose two simple architectural changes to solve this problem. Firstly, we propose drastic thinning of medical imaging CNNs: to significantly
reduce the number of filters per network layer. We show that the U-Net can be dramatically reduced in width and still have sufficient
capacity to model the variability in a popular ultrasound challenge dataset.
Secondly, we suggest the integration of
separable convolutions into the architecture. Separable convolutions offer a more efficient way of parameterising rank deficient convolution
kernels, reducing the number of parameters needed to define each convolutional layer in the network.
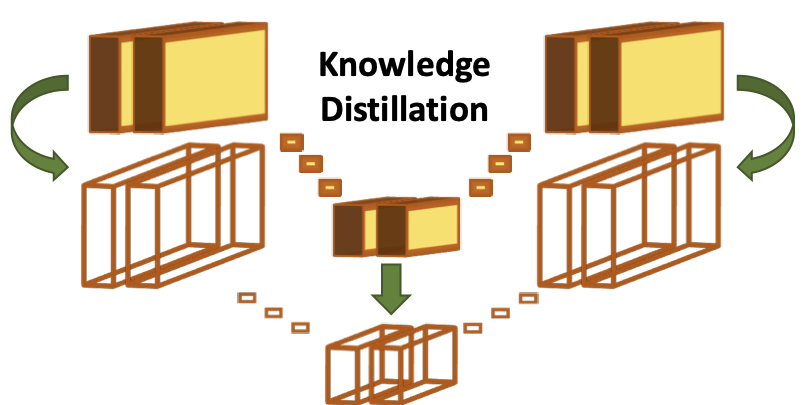
Finally, we propose a novel knowledge distillation technique to boost the performance of the separable convolution models to that of their regular
convolution counterparts, for use in accuracy critical settings. We suggest the incorporation of an additional loss during training, encouraging the
separable convolution model to recreate the intermediate feature maps of its regular convolution 'teacher'.
Outcome
The results in our paper show that our final distilled model has an accuracy statistically equivalent to that of the original U-Net while running
9 times faster, and taking 420 times less space in memory. The model runs at 30 frames per second on a CPU, which is real-time in the context of
ultrasound. It is especially important that real-time inference is demonstrated on CPUs - most CNN inference times are shown on GPUs, which are
expensive and typically an order of magnitude faster at neural network computation.
Finally, we implement our lightweight models on mobile devices, as demonstrated in the video on the left. The video shows a model trained
to segment adipose tissue from fetal ultrasound, operating on a Google Pixel 2.